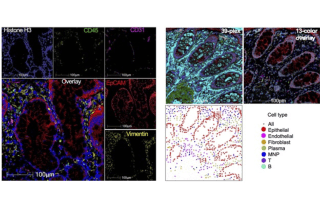
CoLabs has several spatial transcriptomics and proteomics platforms available for UCSF and external investigators.
Getting Started
Combining our expertise in sample processing, staining, imaging, genomics, and data analysis, CoLabs is positioned to provide multi-disciplinary support for spatial profiling projects. Our labs are located near each other to streamline communication and workflows.
While our Spatial Omics instruments are not available for independent use, we will work with you to run your samples. CoLabs will help you with sample preparation, panel design, running slides, and visualization and analysis.
To get started with spatial transcriptomics, please contact our Genomics CoLab Director Walter Eckalbar.
For spatial proteomics, please contact PALM for CellDive and other fluorescence-based technologies.
CoLabs offers whole transcriptome analysis through 10x Genomics CytAssist Visium platform. We also have the ability for subcellular resolution on 10x Genomics Xenium platform, with standard and custom designed panels. Compatible samples include both fresh frozen and fixed tissues. Immunofluorescence and H&E staining can be co-visualized on the same sample section.
| Platform: | CytAssist Visium | Xenium |
|---|---|---|
| Detection: | whole genome |
up to 5,000 genes |
| Resolution: | single cell | sub cellular |
| Sample: | fresh frozen, fixed frozen | fresh frozen, formalin-fixed paraffin-embedded |
| H&E and IF staining: | pre-run | post-run |
To get started with spatial transcriptomics, please contact Genomics CoLab to discuss the overall experimental design and workflow. 10x Genomics provides resources including panel design tools, protocols for sample preparation, and analysis tools.
Once slides are prepared and stained, Genomics CoLab can perform hybridization. However, if you would like to hybridize samples yourself, please coordinate with us to schedule the imaging run as the slides will have limited shelf life. Visualization and analysis are supported through integration among the Disease to Biology, Genomics, PALM, and Data Science CoLabs teams.
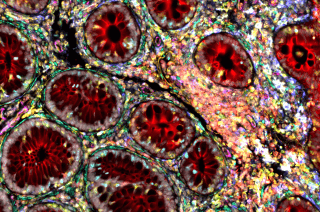
Courtesy of the Kattah Lab
CoLabs offers spatial proteomics analysis using Leica's Cell DIVE Multiplex Imaging system. You provide sample slides, and PALM will perform the staining and imaging. Analysis is also offered by PALM.
| Leica Cell DIVE | |
|---|---|
| Antibody conjugation: | fluorescent dye |
| Panel size: | 40+ panel 22 validated (updated 8/2025) and custom antibody optimization available |
| Sample: | FFPE tissue only, ~4 um thick |
| Compatibility: | H&E |
| Contact: | PALM |
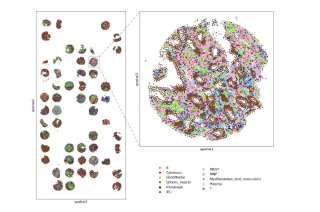
Courtesy of the D2B CoLab
1. Who do I contact to start my experiment? Where are you located?
Spatial transcriptomics:
GenomicsCoLab@ucsf.edu
Medical Sciences Building S813
513 Parnassus Ave (map)Spatial proteomics:
flowassist@ucsf.edu (MIBI) & palm@ucsf.edu (CellDIVE)
Medical Sciences Building S854
513 Parnassus Ave (map)
2. What if I don’t see the approach that I need for my research? Can CoLabs manage instruments purchased by UCSF labs or programs?
CoLabs is enthusiastic about partnering with UCSF investigators and programs to obtain and manage additional equipment that augment our capacity and bring novel capabilities to UCSF research community. Please contact us to discuss options.
3. Can you help with tissue sectioning?
We do not currently offer tissue sectioning services. However, we can provide guidance on sample preparation and recommendations for sectioning and tissue microarray services that we have worked with in the past. Please contact PALM for more information.